GenePlexus
The GenePlexus webserver enables researchers to predict novel genes similar to their genes of interest based on their patterns of connectivity in human genome-scale molecular interaction networks.
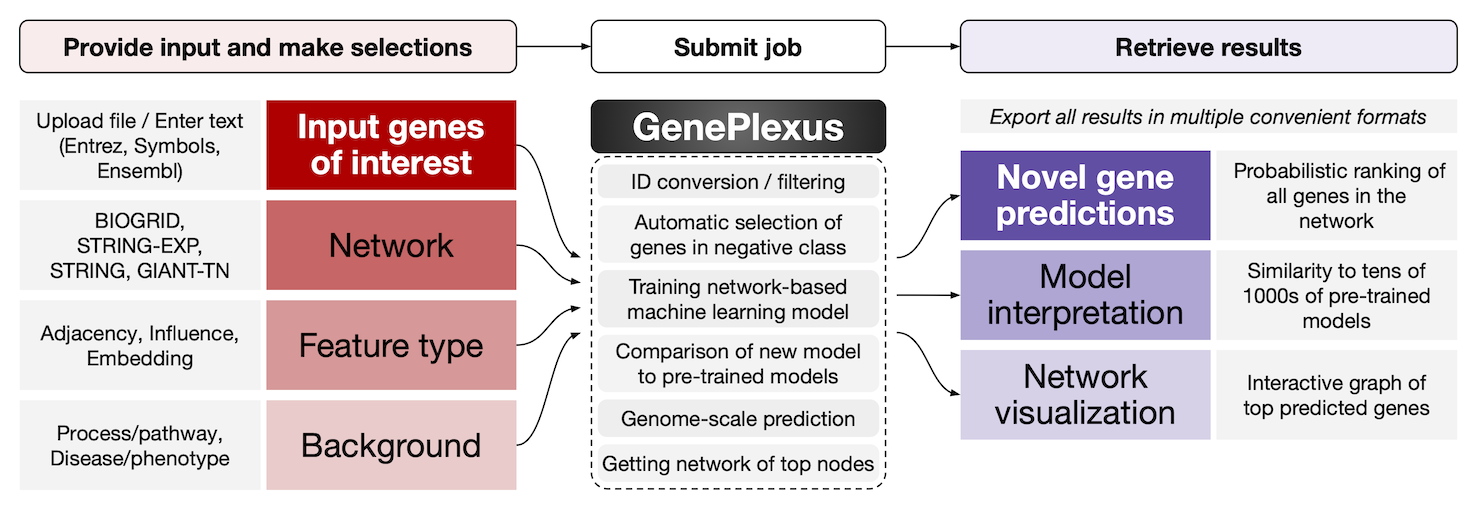
Using GenePlexus
- Click ‘Add Genes’ in the panel on the left to start running GenePlexus on your own genes of interest!
- Watch the GenePlexus tutorial.
- See the About page for an overview of how GenePlexus works.
- See the Help page for detailed help on the various inputs and outputs.
- See an Example run where GenePlexus was used to train a model and predict novel genes associated with the disease (see more below).
- Write to us at help[at]geneplexus.net with feedback on the usability of the web-server and how it helped your research.
- Read the paper:
Mancuso CA, et al. (2022) GenePlexus: a web-server for gene discovery using network-based machine learning, Nuc. Acids Res, doi.org/10.1093/nar/gkac335
About GenePlexus
When a user provides a set of genes to GenePlexus, it trains a custom machine learning (ML) model that captures the patterns of network connectivity of the user’s genes in contrast to other genes in the network. Based on this ML model Geneplexus will:
- Predict other genes in the network that are similar to the input genes based on their network connectivity.
- Help interpret the custom trained ML model by comparing it to pre-trained models for various biological processes and diseases, and
- Provide a visualization of the top predictions in the form of an interactive network graph. All the results can be downloaded/exported in multiple convenient formats.
Job Runtime
While many jobs will only take a few minutes to complete, some networks and feature types require training machine learning models that use 10s of GBs of memory, have more that 25,000 features and use thousands of positive and negative labeled genes. Therefore, some jobs may take up to 10 minutes or more and this time can vary due to latency speeds on the cloud server.
Example
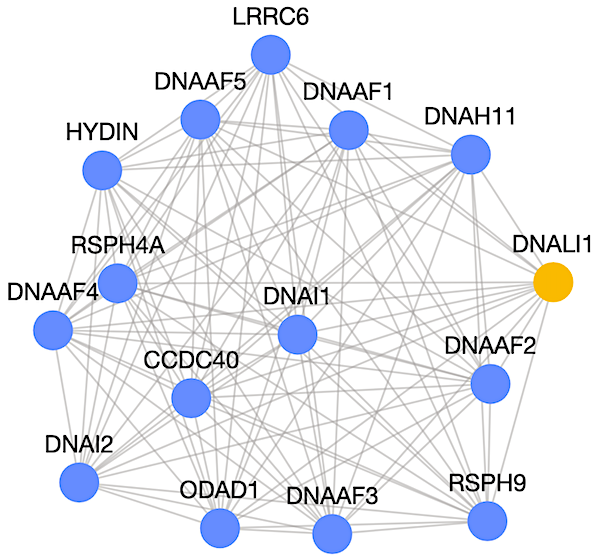
You can visit this page to see the results of an example run where GenePlexus was used to train a model and predict novel genes associated with the disease primary ciliary dyskinesia (PCD).
This run used the adjacency matrix representation of the human STRING network. Since the input genes correspond to a disease, the negative genes were selected based on genes associated with other diseases (from the DisGeNet database). The predicted genes are under the “Gene Predictions” tab. It is notable that the gene DNALI1 (dynein axonemal light intermediate chain 1), which was not included in the original gene list, is strongly predicted to be associated with PCD.
The “Similarities to…” tabs show that the model trained for PCD is similar to those trained for other diseases such as ciliopathy as well as models trained for biological processes that deal with dynein complex and arm assembly.
The "Network Graph" tab shows the network connectivity of up to the top 50 genes based on prediction probability score (an example is seen below where blue nodes are previously known associations and orange nodes are novel predicted associations).
Underlying Model
The network-based approach implemented in GenePlexus has been extensively benchmarked in
Supervised learning is an accurate method for network-based gene classificationR Liu*, CA Mancuso*, A Yannakopoulos, KA Johnson, A Krishnan
Bioinformatics 36 (11), 3457-3465.